DAR help page
This section describe how to use the DAR database website. Statistics about data available in DAR can be found in the Statistics page. Data can be downloaded from the Download page.
The DAR home page - search the database
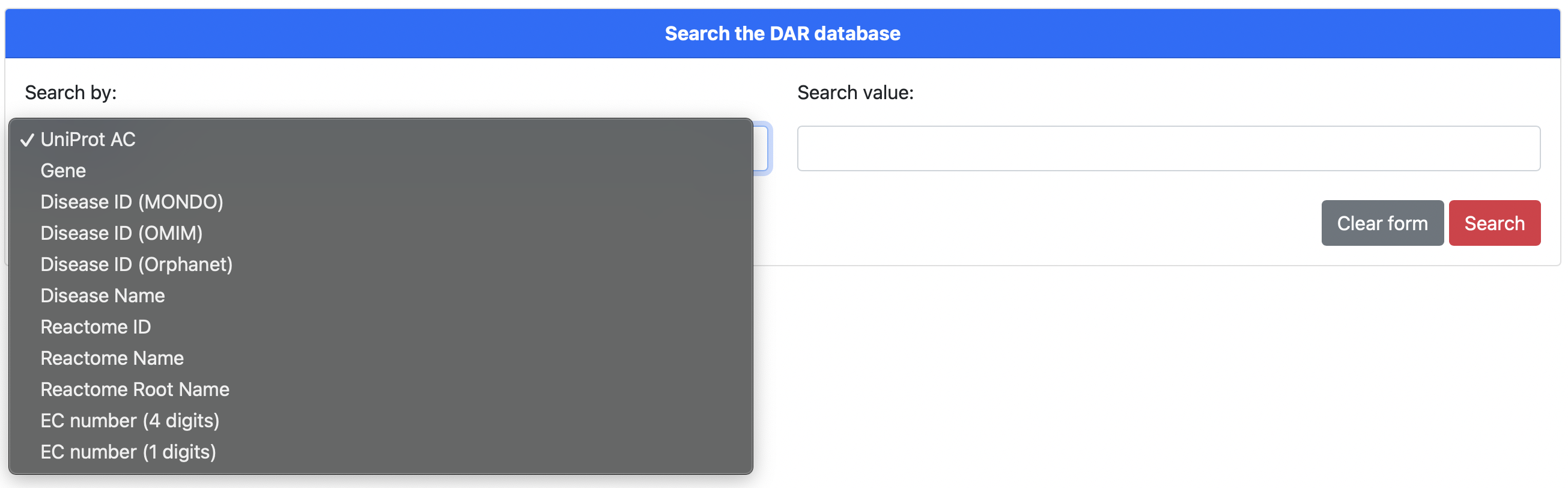
Figure 1. The DAR home page search form
The DAR home page (Figure 1) represents the main access point to the database, allowing the user to make queries by different search fields. In particular, using a selection field, the user can choice to search by the following keys:
- UniProtKB accession
- Gene symbol
- Disease ID (either MONDO, OMIM or Orphanet)
- Disease name
- Reactome pathway ID
- Reactome pathway name
- Reactome root pathway
- EC number (either 4- or 1-digit EC number)
After selecting the search field, the user can specifiy the serach value in the text area. All the queries support autocompletion (Figure 2), to help the user properly selecting the input values. The query can be submitted using the Search button.
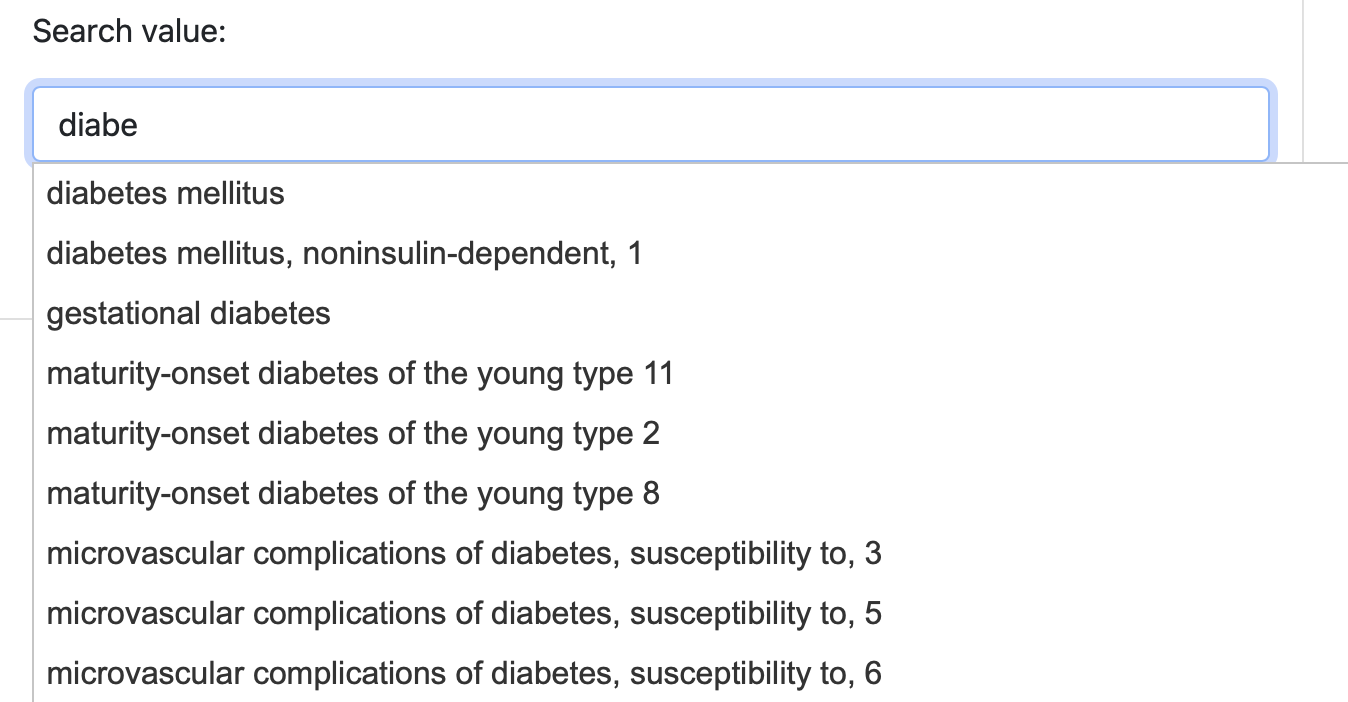
Figure 2. The DAR autocompletion
Browse DAR
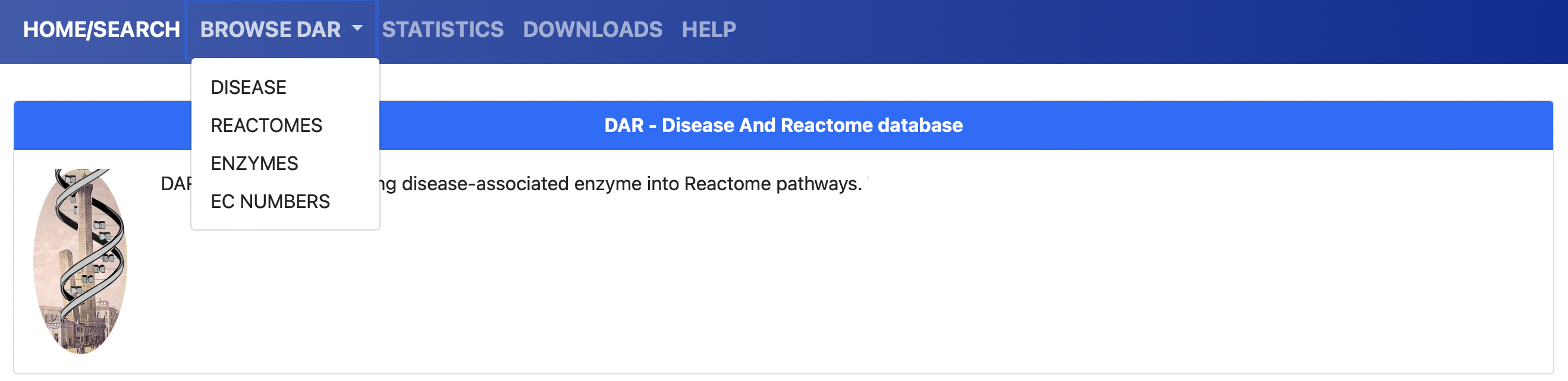
Figure 3. Browse DAR
The user can choose to explore the database content by browsing DAR by different entities. Clicking on the BROWSE entry of the menu on top of the page (Figure 3), the user can choose to brwose DAR by:
- Diseases
- Reactomes
- Enzymes
- EC numbers
Clicking on one of the above browse keys, teh user is redirected to a page containing all entries of the selected type contained in DAR. For instance, in Figure 4 it is shown the Browse by Reactome page. Clicking on the Reactome ID, the user is redirected to the corresponding result page (see next section).

Figure 4. Browse DAR by Reactomes
DAR search results
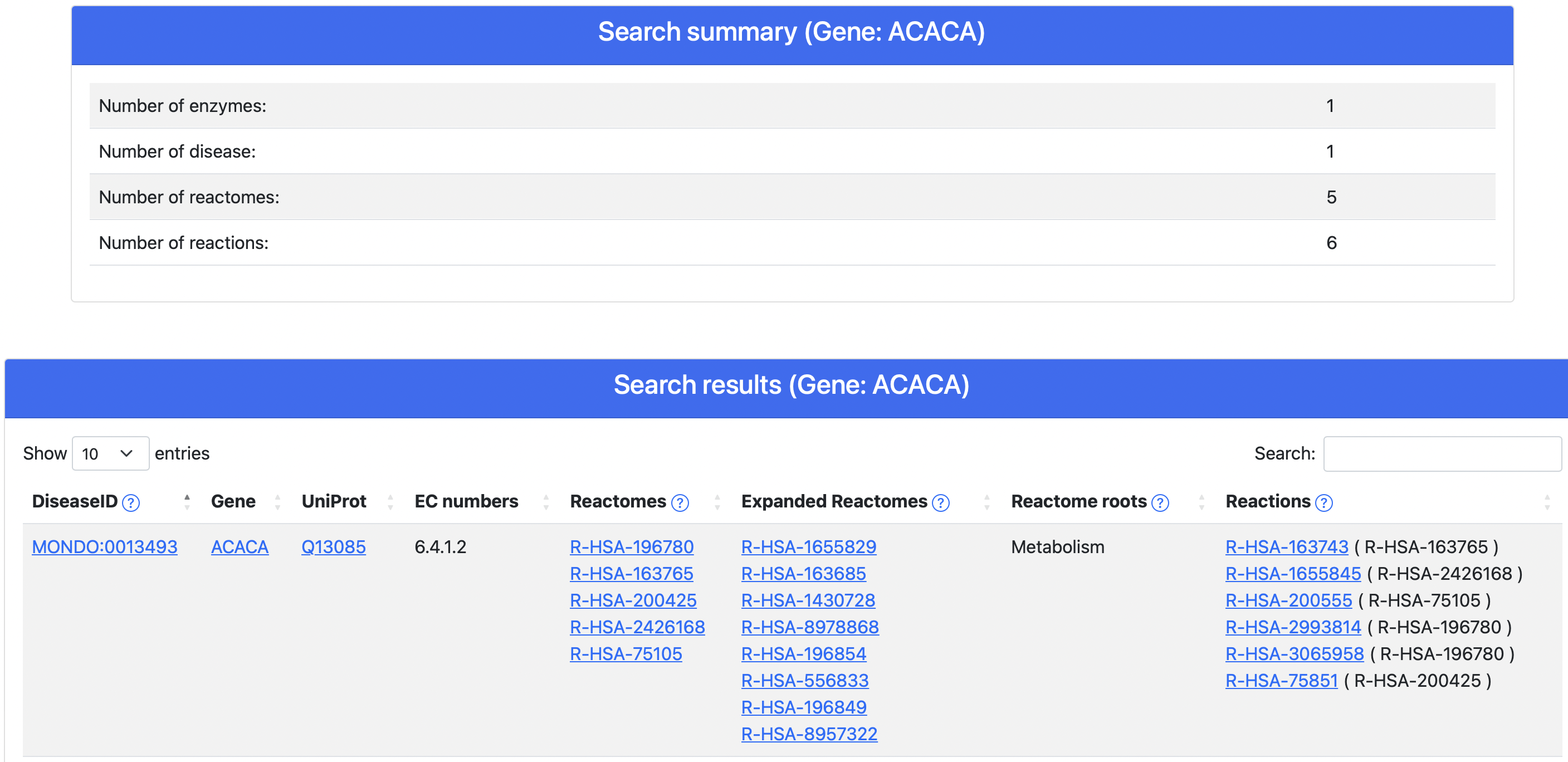
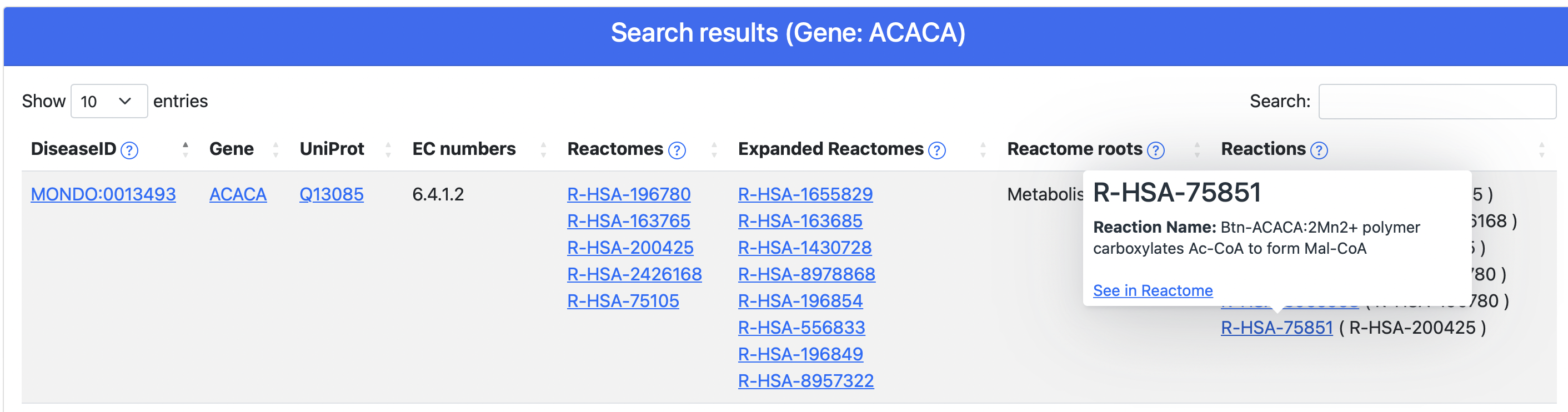
Figure 5. DAR result page
Upon query submission, DAR returns to the user a result page (Figure 5) containing a summary table and the data organized in a tabular format. Each row reports a disease-enzyme association alongside several additional information, including:
- EC numbers
- Reactome pathways (both those directly annotated on the gene/protein as well as those obtained expanding the former through the complete Reactome hierarchical tree)
- Reactome Root names where the annotated pathways map to
- All Reactions (from Reactome) associated to the enzymes
From the table, clicking on the UniProtKB accession or the gene symbol, the user is redirected on the corresponding UniProtKB page. Clicking on disease ID, Reactome IDs or Reactions, a tooltip opens showing details about all these fields as well as links to reference resources:
- Monarch/OMIM/Orphanet for diseases (Figure 6)
- Reactome web site for pathways (Figure 7)
- Reactome web site for reactions (Figure 8)
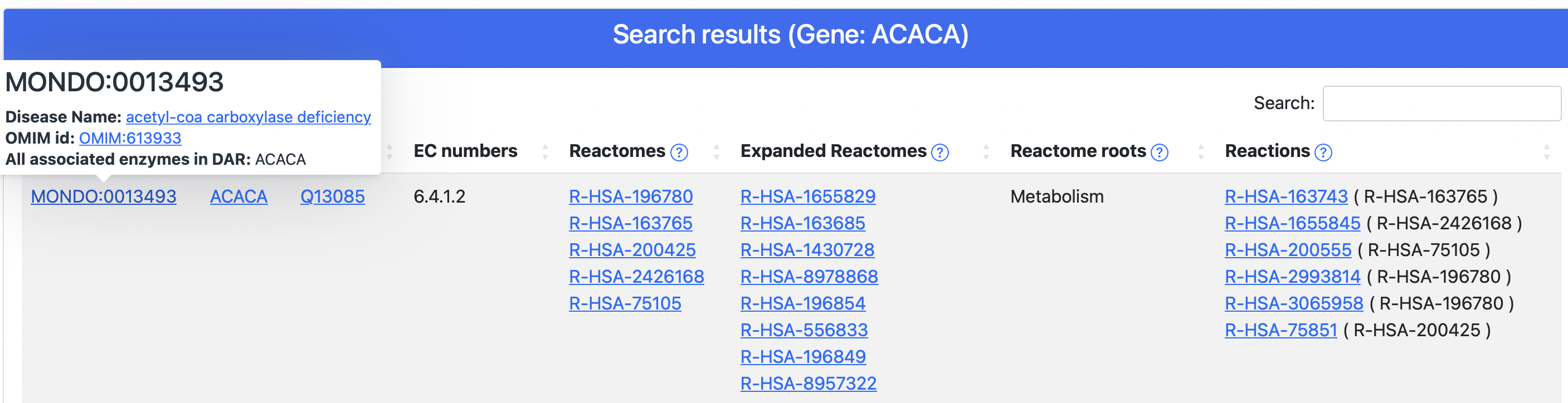
Figure 6. The disease tooltip
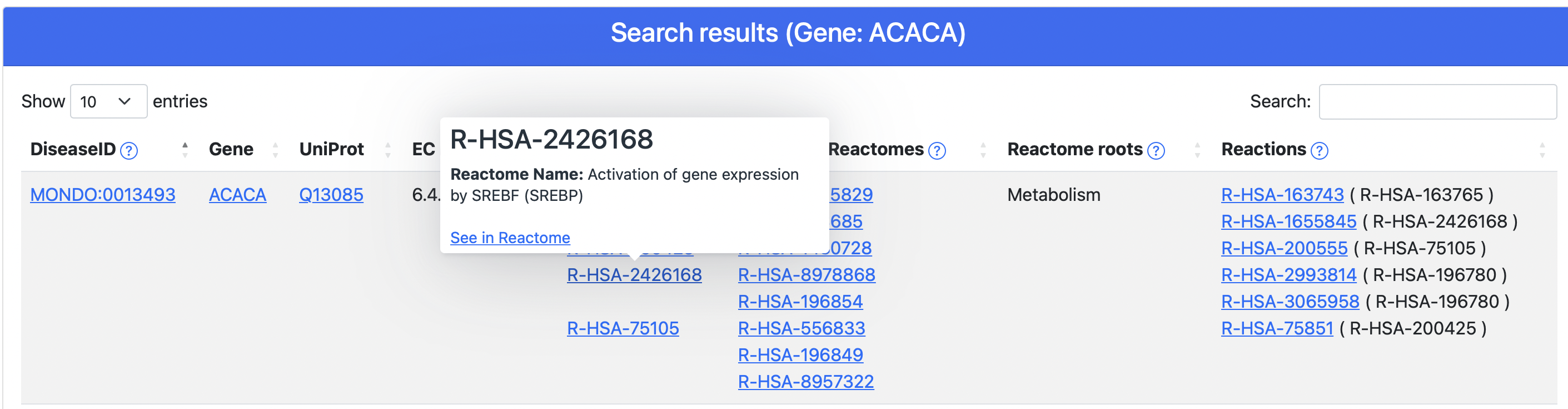
Figure 7. The Reactome tooltip